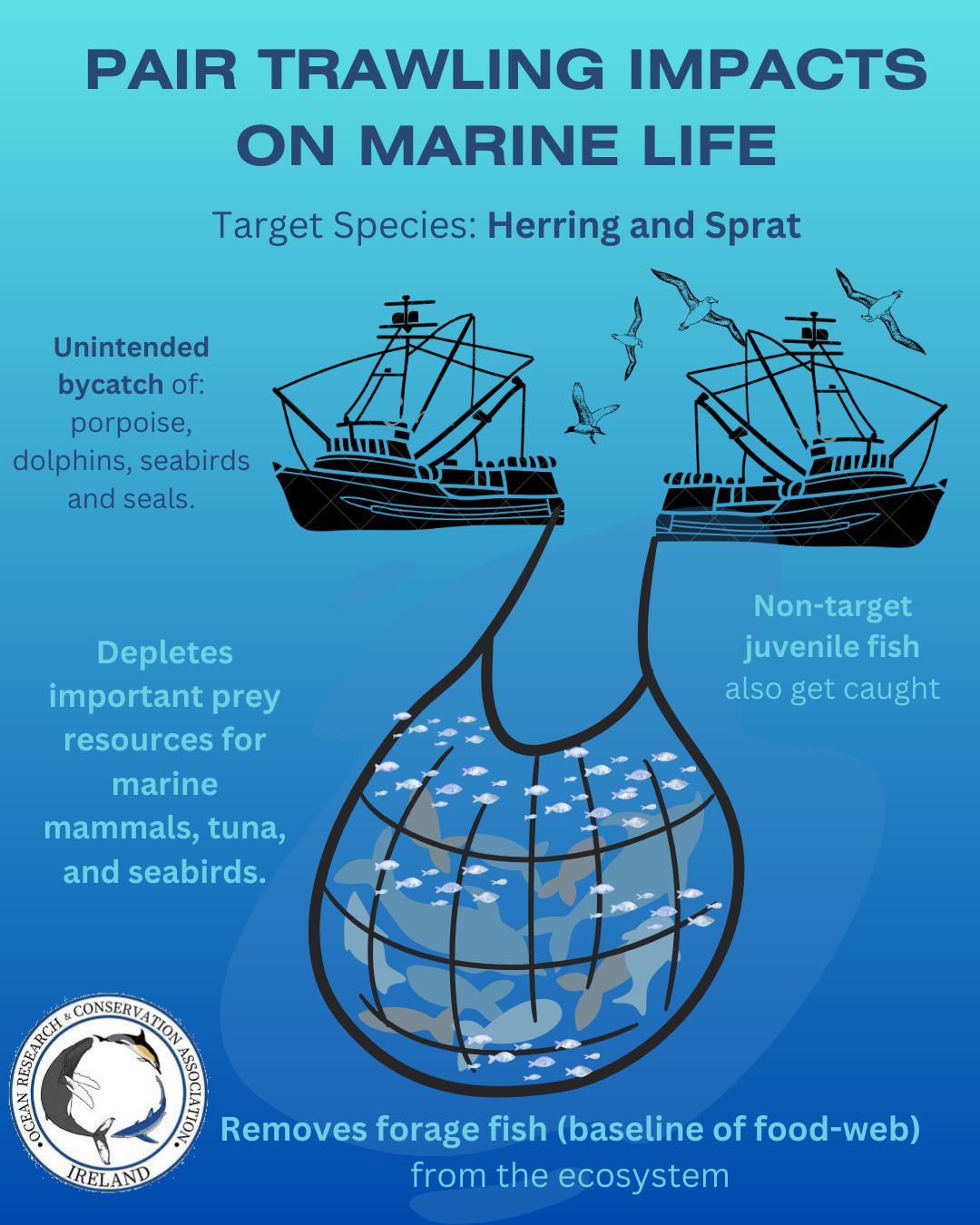
Type D killer whales are the most inbred mammals ever documented.
Type D killer whales are the most inbred mammals ever documented.
ORCA Sci-Comm Team | 28th March, 2023
Killer whales, (Orcinus orca), are the largest members of the dolphin family and one of the most powerful predators on Earth. Populations of killer whales around the world are known for their diverse ecotypes, each exhibiting distinct morphological, behavioral, and ecological characteristics. Although research on killer whales has been extensive, the genomics of type D killer whales have remained little known to date, due to their elusive nature and limited observations. New research has revealed that type D killer whales are the most inbred mammals ever documented. How did this come to be and what are the implications of these findings to their conservation. Read on to find out more.

Killer whales (Orcinus orca) are known to be a polytypic species - meaning they have many genetically distinct subspecies and separate units of geographical populations. Five distinct forms have been described from the Southern Ocean alone, with distinct habitat and prey preferences, in addition to type-specific coloration and body shape and sizes. Demographic-specific characteristics have also been shown in this diverse taxon, for example, the sympatric mammal-eating “transient” (also known as the Bigg’s killer whale) and fish-eating “resident” ecotypes found in the North Pacific are strongly differentiated across the genome, and share ancestry with different outgroup populations. In comparison, the Antarctic sub-groups, types B1, B2, and C, show ecological and morphological differences, but share only partial geographic separation and ancestry, and are only weakly genetically differentiated from one another.
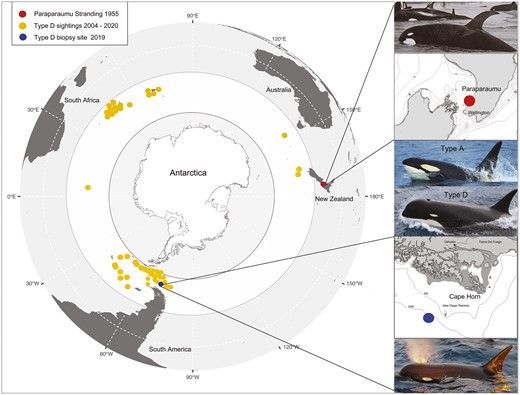
Type D killer whales, which show a preference for offshore waters in the Southern Ocean, have a characteristic bulbous head compared to other killer whale types, and a reduced eyepatch, which is sometimes absent in individuals. The first record of a type D killer whale came from a stranding of an individual from New Zealand 67 years ago, with relatively few observations of the eco/morphotype recorded at sea since. The 48 records of type D killer whales sighted at sea are mostly from observations of depredating animals around fishing vessels) and reports from tourism vessels. Due to challenges faced by field biologists to collect tissue samples of type D killer whales in the field, they have (up until now) evaded scientists hampering the ability to genomic analyses in the lab.
In January 2019, a team of researchers followed reports of type D killer whales that were regularly depredating commercial fishing boats in Chilean waters and encountered a group of approximately 30 individuals. The researchers collected photo ID, and underwater video of the group and collected biopsy samples from 3 individuals.
The new study published in the
Journal of Heredity
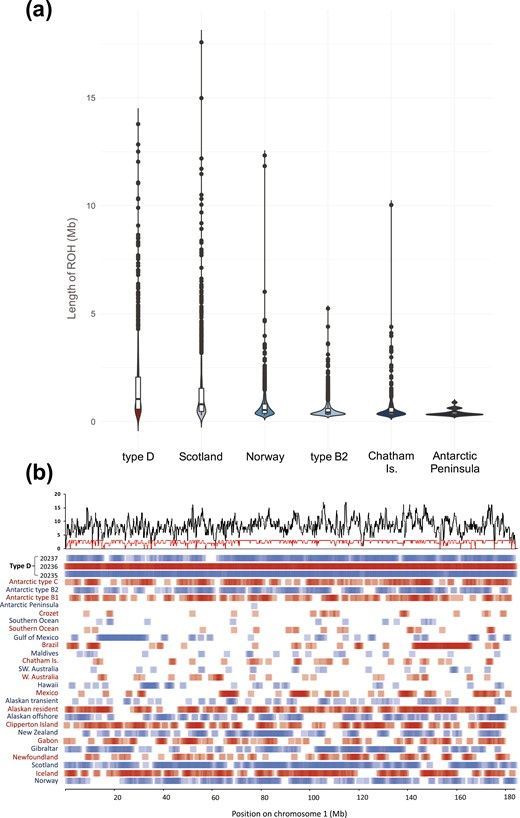
used advanced genomic sequencing techniques and has provided the first genomic characterization of type D killer whales, a unique eco/morphotype with a circumpolar, sub-antarctic distribution. The research revealed that type D effective population size is the lowest estimated from any killer whale genome and indicates a severe population bottleneck, raising concern for the future viability of type D killer whales. As a result, type D killer whale genomes have among the highest levels of inbreeding reported for any mammalian species on Earth! They were also compared to other populations from a global dataset of killer whale genomes and were found to have the highest inbreeding coefficient, higher even than the previously estimated genome from a small Scottish population on the brink of extinction.
They analyzed mitochondrial genomes from the 3 type D killer whales sampled off Chile and found they were all identical, indicating a shared matrilineal genealogy, consistent with the individuals being part of the same matrilineal kin-based social group (typical of killer whales).
The 3 biopsied type D killer whales were all found to be female, based upon the approximate equal mean depth of coverage of the longest autosome and the X-chromosome. These three whales were determined to be a mother and her two full-sibling daughters.
In conclusion, this groundbreaking study provided the first genome sequences of type D killer whales and revealed a shocking revelation, that they are the most extreme example of long-term inbreeding within this species to date.
The extent of the type D killer whale genome was composed by homozygous tracts that exceed those found in some of the most prominent examples of inbreeding in wild populations, e.g. eastern lowland (Gorilla beringei graueri), mountain gorillas (G. b. beringei) and Royale wolves (Canis lupis).
Reference:
Andrew D Foote, Alana Alexander, Lisa T Ballance, Rochelle Constantine, Bárbara Galletti Vernazzani Muñoz, Christophe Guinet, Kelly M Robertson, Mikkel-Holger S Sinding, Mariano Sironi, Paul Tixier, John Totterdell, Jared R Towers, Rebecca Wellard, Robert L Pitman, Phillip A Morin, “Type D” killer whale genomes reveal long-term small population size and low genetic diversity, Journal of Heredity, 2023;, esac070, https://doi.org/10.1093/jhered/esac070
SHARE THIS ARTICLE